


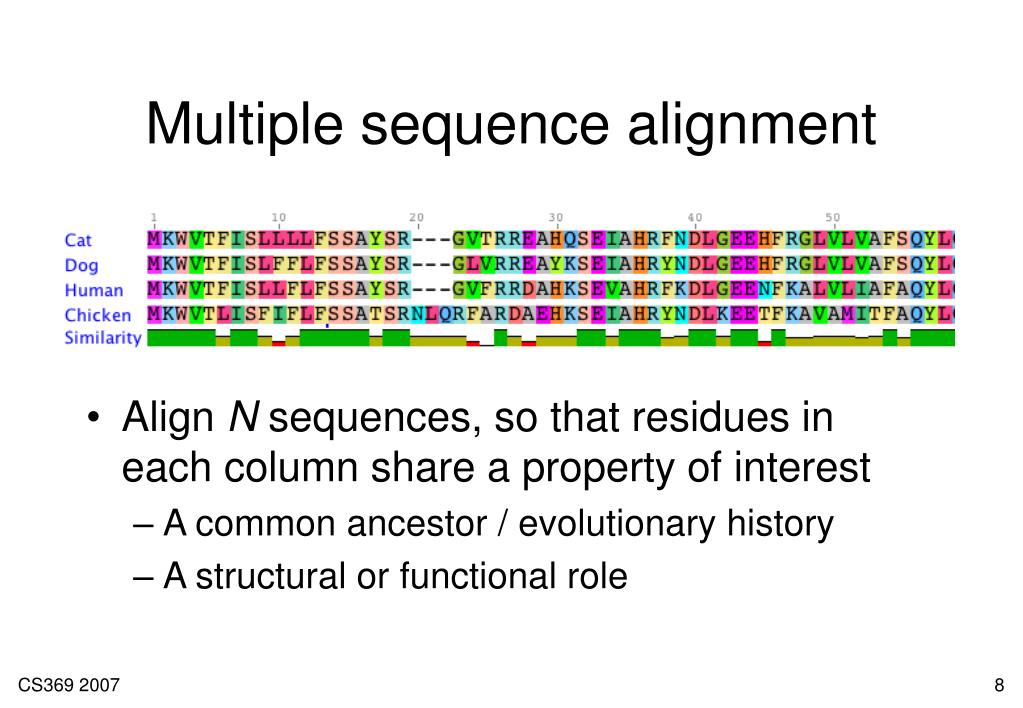
These kinases comprise the Aurora kinases, Polo kinases, and calcium/calmodulin-dependent kinase kinases. A new phylogeny of the protein kinase domains in the human genome based on our alignment indicates that ten kinases previously labeled as “OTHER” can be confidently placed into the CAMK group. The remaining inaccuracy comes from a few structures with shifted elements of secondary structure, and from the boundaries of aligned and unaligned regions, where compromises need to be made to encompass the majority of kinases. From pairwise, all-against-all alignment of 272 human kinase structures, we estimate the accuracy of our MSA to be 97%. The aligned blocks contain well-conserved elements of secondary structure and well-known functional motifs, such as the DFG and HRD motifs. The alignment is arranged in 17 blocks of conserved regions and unaligned blocks in between that contain insertions of varying lengths present in only a subset of kinases. We present a parsimonious, structure-based multiple sequence alignment (MSA) of 497 human protein kinase domains excluding atypical kinases. The key factor in accurate inference by homology is an accurate sequence alignment. You may also open a trace file in the Trace Data Viewer/Editor and send it directly to the Alignment Explorer.Studies on the structures and functions of individual kinases have been used to understand the biological properties of other kinases that do not yet have experimental structures. When you are done, select accessions by press ing the "Fetch" button.Įx 1.2.6: When the status column indicates that all sequences are fetched, press the "Send to Alignment" button to send the fetched sequence data to the Alignment Explorer.Įx 1.2.7: Align the fetched data using the steps detailed in Ex 1.1.2 – Ex 1.1.5.
This will display the Web Fetch dialog window.Įx 1.2.5: Click the box to the left of each accession number whose sequences ’ information you would like to fetch from the web. P ress the "Add to Alignment" button located to the left of the address box. Choose "YES" on the dialog box that appears to indicate that you are creating a DNA sequence.Įx 1.2.2: Activate the Web Explorer tab by selecting Web|Query Gene Banks from the menu.Įx 1.2.3: When the NCBI Entrez site is loaded, select either the nucleotide or protein database, enter a search term into the search box, and press the "GO" button.Įx 1.2.4: When the search results are displayed, select the spe cific search item and choose "Seq uence" from the menu bar. Įx 1.2.1: If the Alignment Explorer already contains sequence data, select the Data | Create new menu command to create a new alignment from Alignment Explorer window.
#Multiple sequence alignment how to
Click "Yes." Now, we will examine how to send sequence data from the Internet (Web Explorer) to the Alignment Explorer. In this case, click "Yes." A final dialog box will appear asking you if you would lik e to open the data file in MEGA.

Another dialog box will appear asking you if the sequence data is protein coding. Enter "HSP 20 Aligned by MEGA" as the title, and click the "OK" button. An input box will appear asking for a title for the data. Enter hsp20_g as the file name, and click the "Save" button. Choose "YES," and then a "Save As" dialog box will appear. A message will appear asking if you would like to save the da ta to a MEGA file. This will allow the current alignment session to be restored for future editing.Įx 1.1.5: Exit the Alignment Explorer by selecting the Data|Exit Alignment Explorer menu item. In order to align sequences contained in a Seque nce Data File, do the following:Įx 1.1.1: A dd unaligned sequences from the hsp20.fas example file into the Alignment Explorer by clicking selecting the Data|Open|Retrieve Sequences from File menu command.Įx 1.1.2: Select the Edit|Select All menu command to select every site for all sequences in the alignment.Įx 1.1.3: Select the Alignment|Align by ClustalW menu command to align the selected sequences data using the ClustalW algorithm.Įx 1.1.4: Save the current alignment session by selecting the Data|Save Session menu item.
#Multiple sequence alignment windows
In this example, we will create an alignment from protein sequence data that will be imported into the alignment editor using different methods.Įx 1.0.1: Start MEGA by double-clicking on the MEGA desktop icon, or by using the Windows start-menu to click on the MEGA icon located in the programs folder.Įx 1.0.2: Launch the Alignment Explorer by selecting the Alignment|Alignment Explorer/CLUSTAL menu command. Creating Multiple Sequence Alignments Creating Multiple Sequence Alignments


 0 kommentar(er)
0 kommentar(er)
